“Quantum computing, with its superior computational capabilities compared to classical approaches, holds the potential to revolutionise numerous scientific domains, including pharmaceuticals,” the team wrote.
Existing classical methods in computational chemistry are not exact – and their cost goes up as the scale of computing grows, the researchers said.
“However, in the current landscape, the involvement of quantum computing in drug discovery is primarily restricted to conceptual validation, with minimal integration into real-world drug design,” the team said.
In answer to that, the team has developed a hybrid quantum computing pipeline targeted for real-world drug discovery, which they were able to validate using two case studies that addressed real problems in drug design.
“Our results demonstrate the potential of a quantum computing pipeline for integration into real world drug design workflows,” the researchers said.
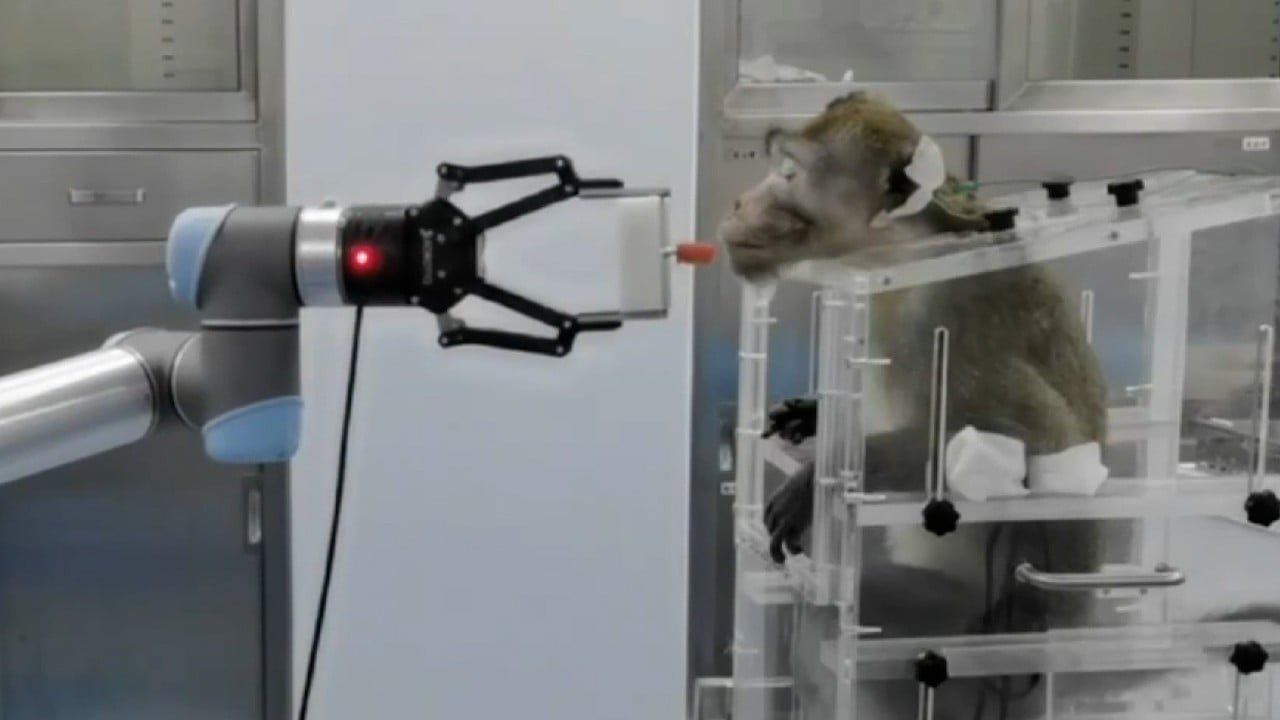
The team sought to carry out two critical tasks in drug discovery: determine the energy needed to cleave or break bonds in a prodrug – a drug that turns from inactive to active inside the body – and the simulation of covalent bonds, a chemical bond where atoms share electrons.
One strategy to activate these drugs is the breaking of carbon-carbon bonds. According to the team, the calculation of an energy barrier for cleavage of these bonds is “crucial”, as it determines whether it can happen spontaneously within the body.
They compared their computing results with a paper from 2022 that used classical computing methods to determine the energy barrier alongside laboratory experimentation.
Analysis using the quantum computer agreed with the previous study, with both analyses determining the drug could undergo a spontaneous reaction within biological organisms.
“Our results demonstrate the effectiveness of quantum computing … as well as the versatility and plug-and-play advantages of our pipeline,” the researchers wrote.
In their second case study, the team sought to determine the activity of another anticancer drug, sotorasib, known as a KRAS (Kirsten Rat Sarcoma) inhibitor, which inhibits a specific KRAS gene mutation, G12C.
Finding medication for mutations of this oncogene has been a challenge, as it needs to form a covalent bond with the target in order to inhibit it.
Quantum mechanics and molecular mechanics simulations – vital simulations in post-design drug validation – were used to examine the drug target interaction. The team used a hybrid computing method, meaning they started with a quantum emulator before moving to a quantum computer.
After performing hybrid quantum computing validation on sotorasib and the target mutation, the team observed that a strong covalent bond formed between them – which could offer insight into the drug’s efficacy.
“This understanding is pivotal for the rational design of future inhibitors targeting similar mutations,” the researchers said, adding that it would underpin future advancement of the speed and accuracy of drug discovery using quantum computing.
“In this study, we have established a model pipeline that enables quantum computers to tackle real-world drug discovery problems,” they said.
“The universality of our pipeline highlights its potential as a foundational tool, empowering researchers with a ready-to-use computational resource.”
They said even drug design experts without a background in quantum computing would be able to use it.
They also said more work was needed to improve the accuracy of quantum computing methods for drug discovery. One challenge is the current limitations of quantum computers, such as longer computational time and errors.



Hi Lucofi Owner,
My name is Eric and I’m betting you’d like your website Lucofi to generate more leads.
Here’s how:
LeadConnect is a software widget that works on your site, ready to capture any visitor’s Name, Email address, and Phone Number. It signals you as soon as they say they’re interested – so that you can talk to that lead while they’re still there at Lucofi.
https://resultleadgeneration.com for a live demo now.
Plus, now that you’ve got their phone number, our new SMS Text With Lead feature enables you to start a text (SMS) conversation – answer questions, provide more info, and close a deal that way.
If they don’t take you up on your offer then, just follow up with text messages for new offers, content links, even just “how you doing?” notes to build a relationship.
https://resultleadgeneration.com to discover what LeadConnect can do for your business.
The difference between contacting someone within 5 minutes versus a half-hour means you could be converting up to 100X more leads today!
Try LeadConnect and get more leads now.
Eric
PS: The studies show 7 out of 10 visitors don’t hang around – you can’t afford to lose them!
LeadConnect offers a complimentary 14-day trial – and it even includes International Long Distance Calling.
You have customers waiting to talk with you right now… don’t keep them waiting.
https://resultleadgeneration.com to try LeadConnect now.
If you’d like to Want to receive fewer emails, or none whatsoever? Update your email preferences by visiting https://resultleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hello to the Lucofi Owner,
I am Eric, and unlike many emails you may receive, I would like to share a note of positive feedback – well done!
What for?
Part of my role is to examine websites, and the work you have done with Lucofi certainly stands out.
It is clear you have taken building a website seriously and invested real effort into developing something of quality.
However, there is a question…
When someone like me finds your site – maybe at the top of the search results (good job, by the way) or through a link, how can you tell?
More importantly, how can you connect with that visitor?
Research indicates that many visitors leave quickly.
Here is a way to create immediate engagement that might be new to you:
Web Visitors Into Leads is a tool that operates on your site, ready to gather each visitor’s name, email address, and phone number. It alerts you promptly when they are interested – so you can speak with them while they are viewing Lucofi.
Please visit https://actionleadgeneration.com to view a live demonstration of Web Visitors Into Leads today and see exactly how it operates.
It can be very helpful for your business – and it gets even better… after you have their phone number, with our text messaging feature, you can begin a conversation promptly (there’s a significant difference between connecting within a few minutes compared to waiting much longer).
Additionally, even if you do not reach a mutual understanding at once, you can maintain contact later with text messages for additional resources, content links, or follow-ups to build a rapport.
Everything described is straightforward, convenient, and effective.
Visit https://actionleadgeneration.com to learn what Web Visitors Into Leads can provide for your business.
You could be engaging with significantly more potential contacts soon!
Eric
P.S. Web Visitors Into Leads includes a 14-day evaluation period and supports international communication. There are individuals ready to speak with you now, so please do not keep them waiting.
Visit https://actionleadgeneration.com to explore Web Visitors Into Leads today.
If you’d like to Want to receive fewer emails, or none whatsoever? Update your email preferences by visiting https://actionleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hello Lucofi Owner!
My name is Eric and I’m betting you’d like your website Lucofi to generate more leads.
Here’s how:
Web Visitors Into Leads is a software widget that works on your site, ready to capture any visitor’s Name, Email address, and Phone Number. It signals you as soon as they say they’re interested – so that you can talk to that lead while they’re still there at Lucofi.
https://boltleadgeneration.com for a live demo now.
Plus, now that you’ve got their phone number, with our new SMS Text With Lead feature, you can automatically start a text (SMS) conversation – answer questions, provide more info, and close a deal that way.
If they don’t take you up on your offer then, just follow up with text messages for new offers, content links, even just “how you doing?” notes to build a relationship.
https://boltleadgeneration.com to discover what Web Visitors Into Leads can do for your business.
The difference between contacting someone within 5 minutes versus a half-hour means you could be converting up to 100X more leads today!
Try Web Visitors Into Leads and get more leads now.
Eric
PS: The studies show 7 out of 10 visitors don’t hang around – you can’t afford to lose them!
Web Visitors Into Leads offers a complimentary 14-day trial – and it even includes International Long Distance Calling.
You have customers waiting to talk with you right now… don’t keep them waiting.
https://boltleadgeneration.com to try Web Visitors Into Leads now.
If you’d like to Want to receive fewer emails, or none whatsoever? Update your email preferences by visiting https://boltleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hello to the Lucofi Owner,
I am Eric and I recently came across your website at Lucofi. I found it after a search, so your visibility seems to be working well.
Your content looks helpful.
However, one thing might be missing:
A simple, prompt way for visitors to connect with you.
Research suggests that many visitors leave a site quickly without providing any contact information. Once they go, there’s no way to follow up.
Consider this approach:
Web Visitors Into Leads is a tool that operates on your site, ready to gather each visitor’s name, email address, and phone number. You will know as soon as someone shows interest, allowing you to speak with them while they’re still viewing your site.
View a Demonstration https://actionleadgeneration.com
Once you have their phone number, you can also begin a text conversation. If they aren’t ready to engage at that exact moment, you can maintain communication later by sharing additional updates or simply checking in.
Everything described is straightforward to implement and may help improve your interactions with potential contacts.
Please visit https://actionleadgeneration.com to learn more about what Web Visitors Into Leads can do for you. It could help you connect with more interested individuals right now.
Eric
P.S. Web Visitors Into Leads includes a trial period and supports international communication. There may be individuals ready to speak with you now, so please don’t miss the opportunity.
Visit https://actionleadgeneration.com to learn more.
If you’d like to Want to receive fewer emails, or none whatsoever? Update your email preferences by visiting https://actionleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hello, Eric here with a quick thought about your website Lucofi
Cool website!
My name’s Eric, and I just found your site – Lucofi – while surfing the net. You showed up at the top of the search results, so I checked you out. Looks like what you’re doing is pretty cool.
But if you don’t mind me asking – after someone like me stumbles across Lucofi, what usually happens?
Is your site generating leads for your business?
I’m guessing some, but I also bet you’d like more… studies show that 7 out of 10 who land on a site wind up leaving without a trace.
Not good.
Here’s a thought…
How about making it really EASY for every visitor who shows up to get a personal phone call from you as soon as they hit your site…
You can –
Web Visitor is a software widget that works on your site, ready to capture any visitor’s Name, Email address, and Phone Number. It lets you know IMMEDIATELY – so that you can talk to that lead while they’re literally looking over your site.
https://resultleadgeneration.com to try out a Live Demo with Web Visitor now to see exactly how it works.
You’ll be amazed—the difference between contacting someone within 5 minutes versus a half-hour or more later could increase your results 100-fold.
It gets even better… once you’ve captured their phone number, with our new SMS Text With Lead feature, you can automatically start a text (SMS) conversation – immediately… and contacting someone in that 5-minute window is 100 times more powerful than reaching out 30 minutes or more later.
Plus, with text messaging you can follow up later with new offers, content links, even just follow-up notes to keep the conversation going.
Everything I’ve just described is simple, easy, and effective.
https://resultleadgeneration.com to discover what Web Visitor can do for your business, potentially converting up to 100X more eyeballs into leads today!
Eric
PS: Web Visitor offers a complimentary 14-day trial – and it even includes International Long Distance Calling.
You have customers waiting to talk with you right now… don’t keep them waiting.
https://resultleadgeneration.com to try Web Visitor now.
If you’d like to Want to receive fewer emails, or none whatsoever? Update your email preferences by visiting https://resultleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hello Lucofi Owner!
Eric here with a quick thought about your website Lucofi…
I’m on the internet a lot and I look at a lot of business websites.
Like yours, many of them have great content.
But all too often, they come up short when it comes to engaging and connecting with anyone who visits.
I get it – it’s hard. Studies show 7 out of 10 people who land on a site abandon it in moments without leaving even a trace. You got the eyeball, but nothing else.
Here’s a solution for you…
LeadConnect is a software widget that works on your site, ready to capture any visitor’s Name, Email address, and Phone Number. You’ll know immediately they’re interested, and you can call them directly to talk with them while they’re literally looking over your site.
Visit https://blastleadgeneration.com to try out a Live Demo with LeadConnect now to see exactly how it works.
It could be huge for your business – and because you’ve got that phone number, with our new SMS Text With Lead feature, you can automatically start a text (SMS) conversation immediately… and contacting someone in that 5-minute window is 100 times more powerful than reaching out 30 minutes or more later.
Plus, with text messaging, you can follow up later with new offers, content links, even just follow-up notes to keep the conversation going.
Everything I’ve just described is extremely simple to implement, cost-effective, and profitable.
Visit https://blastleadgeneration.com to discover what LeadConnect can do for your business.
You could be converting up to 100X more leads today!
Eric
PS: LeadConnect offers a complimentary 14-day trial – you could be converting up to 100x more leads immediately!
It even includes International Long Distance Calling.
You have customers waiting to talk with you right now… don’t keep them waiting.
Visit https://blastleadgeneration.com to try LeadConnect now.
If you’d like to Want to receive fewer emails, or none whatsoever? Update your email preferences by visiting https://blastleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hello to the Lucofi Owner. This is Eric and I noticed Lucofi a short time ago.
It appears well structured… now what?
What I mean is, when someone like me discovers your website – either through a search or just browsing around – what happens next? Are you receiving meaningful inquiries from your site, or enough to meet your needs?
Honestly, most business websites come up a bit short when it comes to attracting engaged visitors. Research indicates that 70% of a site’s visitors leave and never return after only a moment.
Here is an idea…
What about making it simple for each visitor who arrives to receive a direct phone call from you as soon as they reach your site…
This is possible –
Web Visitors Into Leads is a tool that operates on your site, designed to collect each visitor’s name, email address, and phone number. It notifies you the moment they indicate interest – so that you can speak to that person while they are browsing your site.
Please visit https://actionleadgeneration.com to view a live demonstration of Web Visitors Into Leads and understand how it operates.
You will notice a significant difference between connecting with someone within 5 minutes compared to a half-hour or more later. This can greatly improve the likelihood of meaningful engagement.
It becomes even more helpful. After you have their phone number, with our SMS text communication feature, you can promptly begin a text conversation.
That way, even if you do not reach a conclusion immediately, you can stay connected through text messages for informational updates, content links, or simple check-in notes to establish a relationship.
Quite helpful.
Please visit https://actionleadgeneration.com to learn how Web Visitors Into Leads can support your business.
You can potentially connect with many more interested individuals as soon as possible.
Eric
PS: Web Visitors Into Leads provides a no-cost 14-day evaluation period. It also supports international communication. There are interested individuals ready to speak with you now, so please do not delay.
Please visit https://actionleadgeneration.com to explore Web Visitors Into Leads.
If you’d like to Want to receive fewer emails, or none whatsoever? Update your email preferences by visiting https://actionleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hello Lucofi Owner,
This is Eric and for just a second, imagine this:
– Someone does a search and ends up at Lucofi.
– They hang out for a moment to check it out. I’m interested… but… maybe…
– And then they click the back button and check out the other search results instead.
– Bottom line – you got an eyeball, but nothing else.
– They’re gone.
This isn’t really your fault – it happens a lot – studies show 7 out of 10 visitors to any site leave without leaving a trace.
But you can fix that.
Web Visitors Into Leads is a software that operates on your site, ready to capture any visitor’s Name, Email address, and Phone Number. You will know instantly they’re interested and you can call them directly to chat with them – literally while they’re still on the internet looking at your site.
Please see this URL to test a Live Demo with Web Visitors Into Leads now to see exactly how it works and even give it a try… it could be huge for your business: https://trustedleadgeneration.com
Time is of the essence when it comes to connecting with leads – the difference between contacting someone within 5 minutes compared to 30 minutes later can be significant.
Moreover, now that you’ve got that phone number, with our new SMS Text With Lead feature, you can instantly start a text (SMS) conversation immediately… which is so effective, because connecting with someone within the first 5 minutes is 100 times more efficient than waiting 30 minutes or more later.
The new text messaging feature lets you follow up frequently with new offers, content links, even just follow-up notes to develop a relationship.
Everything I’ve just described is very easy to implement, cost-effective, and beneficial.
Please see this URL to discover what Web Visitors Into Leads can do for your business, possibly converting up to 100X more visitors into leads today: https://trustedleadgeneration.com
Eric
PS: Web Visitors Into Leads offers a free 14-day trial – and it even includes International Long Distance Calling.
You have clients waiting to speak with you right now… don’t keep them waiting.
Please see this URL to try Web Visitors Into Leads now: https://trustedleadgeneration.com
If you’d like to Want to receive less emails, or none whatsoever? Update your email preferences by clicking here. https://trustedleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hello Lucofi Owner,
My name is Eric and unlike a lot of emails you might get, I wanted to instead provide you with a word of encouragement – Congratulations
What for?
Part of my job is to check out websites and the work you’ve done with Lucofi Admin definitely stands out.
It’s clear you took building a website seriously and made a real investment of time and resources into making it top quality.
There is, however, a catch… more accurately, a question…
So when someone like me happens to find your site – maybe at the top of the search results (nice job BTW) or just through a random link, how do you know?
More importantly, how do you make a connection with that person?
Studies show that 7 out of 10 visitors don’t stick around – they’re there one second and then gone with the wind.
Here’s a way to create INSTANT engagement that you may not have known about…
Web Visitor is a software widget that works on your site, ready to capture any visitor’s Name, Email address, and Phone Number. It lets you know INSTANTLY that they’re interested – so that you can talk to that lead while they’re literally checking out Lucofi.
https://resultleadgeneration.com to try out a Live Demo with Web Visitor now to see exactly how it works.
It could be a game-changer for your business – and it gets even better… once you’ve captured their phone number, with our new SMS Text With Lead feature, you can automatically start a text (SMS) conversation – immediately (and there’s literally a 100X difference between contacting someone within 5 minutes versus 30 minutes.)
Plus then, even if you don’t close a deal right away, you can connect later on with text messages for new offers, content links, even just follow up notes to build a relationship.
Everything I’ve just described is simple, easy, and effective.
https://resultleadgeneration.com to discover what Web Visitor can do for your business.
You could be converting up to 100X more leads today!
Eric
PS: Web Visitor offers a complimentary 14-day trial – and it even includes International Long Distance Calling.
You have customers waiting to talk with you right now… don’t keep them waiting.
https://resultleadgeneration.com to try Web Visitor now.
If you’d like to Want to receive fewer emails, or none whatsoever? Update your email preferences by visiting https://resultleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hello to the Lucofi Owner,
I am Eric, and for a moment, consider this scenario:
– Someone searches online and arrives at Lucofi.
– They spend a short time reviewing your site. They might be curious, but not fully decided.
– Then, they leave and look at other options.
– In the end, you received a visitor, but no meaningful connection.
This situation occurs often. It’s very common for most site visitors to leave without taking further action.
You can address this:
Web Visitors Into Leads is a resource that operates on your site, ready to gather each visitor’s name, email address, and phone number. It will alert you right when they show interest, allowing you to connect with them while they are still exploring your site.
View a Demonstration https://actionleadgeneration.com
Prompt contact makes a difference. Engaging with someone shortly after they visit is far more effective than waiting until much later.
Moreover, once you have their phone number, you can begin a text-based conversation. If they are not ready to move forward at that exact moment, you can maintain contact by sharing helpful information later on.
Everything described is straightforward and practical.
Please visit the link above to see how Web Visitors Into Leads can benefit your business. You may find yourself connecting with significantly more interested individuals soon.
Eric
P.S. Web Visitors Into Leads provides a no-cost evaluation period. It also supports various communication methods. There may be potential clients interested in speaking with you right now, so don’t let them slip away.
Please visit https://actionleadgeneration.com to learn more about Web Visitors Into Leads.
If you’d like to Want to receive fewer emails, or none whatsoever? Update your email preferences by visiting https://actionleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hello Lucofi Owner,
My name’s Eric and I just found your site Lucofi
It’s got a lot going for it, but here’s an idea to make it even MORE effective.
Visit https://resultleadgeneration.com for a live demo now.
Web Visitor is a software widget that works on your site, ready to capture any visitor’s Name, Email address, and Phone Number. It signals you the moment they let you know they’re interested – so that you can talk to that lead while they’re literally looking over your site.
https://resultleadgeneration.com to try out a Live Demo with Web Visitor now to see exactly how it works.
It could be huge for your business – and because you’ve got that phone number, with our new SMS Text With Lead feature, you can automatically start a text (SMS) conversation – immediately… and contacting someone in that 5-minute window is 100 times more powerful than reaching out 30 minutes or more later.
Plus, with text messaging you can follow up later with new offers, content links, even just follow-up notes to keep the conversation going.
Everything I’ve just described is simple, easy, and effective.
https://resultleadgeneration.com to discover what Web Visitor can do for your business.
You could be converting up to 100X more leads today!
Eric
PS: Studies show that 70% of a site’s visitors disappear and are gone forever after just a moment. Don’t keep losing them.
Web Visitor offers a complimentary 14-day trial – and it even includes International Long Distance Calling.
You have customers waiting to talk with you right now… don’t keep them waiting.
https://resultleadgeneration.com to try Web Visitor now.
If you’d like to Want to receive fewer emails, or none whatsoever? Update your email preferences by visiting https://resultleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hello Lucofi Owner
My name’s Eric and I’m betting you’d like your website Lucofi to generate more leads.
Here’s how:
Web Visitors Into Leads is a software widget that works on your site, ready to capture any visitor’s Name, Email address, and Phone Number. It signals you as soon as they say they’re interested – so that you can talk to that lead while they’re still there at Lucofi.
https://resultleadgeneration.com for a live demo now.
And now that you’ve got their phone number, our new SMS Text With Lead feature enables you to start a text (SMS) conversation – answer questions, provide more info, and close a deal that way.
If they don’t take you up on your offer then, just follow up with text messages for new offers, content links, even just “how are you doing?” notes to build a relationship.
https://resultleadgeneration.com to discover what Web Visitors Into Leads can do for your business.
The difference between contacting someone within 5 minutes versus a half-hour means you could be converting up to 100X more leads today!
Try Web Visitors Into Leads and get more leads now.
Eric
PS: Studies show that 70% of a site’s visitors disappear and are gone forever after just a moment. Don’t keep losing them.
Web Visitors Into Leads offers a complimentary 14-day trial – and it even includes International Long Distance Calling.
You have customers waiting to talk with you right now… don’t keep them waiting.
https://resultleadgeneration.com to try Web Visitors Into Leads now.
If you’d like to Want to receive fewer emails, or none whatsoever? Update your email preferences by visiting https://resultleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hello Lucofi,
I’m Eric and I recently discovered your website – Lucofi – in the search results.
This is what that means to me…
Your SEO’s working.
You’re receiving eyeballs – at least mine.
Your content’s pretty good, would not change a thing.
BUT…
Eyeballs do not pay the bills.
CUSTOMERS do pay the bills.
And studies show that 7 out of 10 visitors like Lucofi will drop by, have a look, and then leave without doing anything else.
It’s like they never even were there.
You can fix this.
You can make it extremely easy for them to show interest, say, okay, let’s talk without needing them to even pull their phone from their pocket… thanks to Web Visitors Into Leads.
Web Visitors Into Leads is a software widget that sits on your website, ready and waiting to capture any visitor’s Name, Email address, and Phone Number. It allows you know immediately – so you can speak with that lead immediately… before they leave.
Please visit the following URL to experience a Live Demo with Web Visitors Into Leads now to see precisely how it works and even give it a try… it could be huge for your business: https://trustedleadgeneration.com
Time is of the essence when it comes to connecting with leads – the difference between contacting someone within 5 minutes compared to 30 minutes implies you can be converting up to 100X more leads now!
Moreover, now that you’ve got their phone number, with our new SMS Text With Lead feature, you can instantly start a text (SMS) conversation immediately… which is so effective, because connecting with someone within the first 5 minutes is 100 times more efficient than waiting 30 minutes or more later.
The new text messaging feature lets you follow up frequently with new offers, content links, even just follow-up notes to develop a relationship.
Everything I’ve just described is very easy to implement, cost-effective, and beneficial.
Please visit the following URL to discover what Web Visitors Into Leads can do for your business, possibly converting up to 100X more visitors into leads today: https://trustedleadgeneration.com
Eric
PS: Web Visitors Into Leads offers a free 14 days trial!
It even includes International Long Distance Calling.
Paying customers are waiting.
Start connecting today by visiting https://trustedleadgeneration.com to try Web Visitors Into Leads now.
If you’d like to Want to receive less emails, or none whatsoever? Update your email preferences by clicking here.
https://trustedleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hello Lucofi Owner,
My name is Eric and I just came across your website at Lucofi…
Looks great… but now what?
By that I mean, when someone like me finds your website – either through Search or just bouncing around – what happens next? Do you get a lot of leads from your site, or at least enough to make you happy?
Honestly, most business websites fall a bit short when it comes to generating paying customers. Studies show that 70% of a site’s visitors disappear and are gone forever after just a moment.
Here’s an idea…
How about making it really EASY for every visitor who shows up to get a personal phone call from you as soon as they hit your site…
You can –
Web Visitors Into Leads is a software widget that works on your site, ready to capture any visitor’s Name, Email address, and Phone Number. It signals you as soon as they let you know they’re interested – so that you can talk to that lead while they’re literally looking over your site.
https://resultleadgeneration.com to try out a Live Demo with Web Visitors Into Leads now to see exactly how it works.
You’ll be amazed—the difference between contacting someone within 5 minutes versus a half-hour or more later could increase your results 100-fold.
It gets even better… once you’ve captured their phone number, with our new SMS Text With Lead feature, you can automatically start a text (SMS) conversation.
That way, even if you don’t close a deal right away, you can follow up with text messages for new offers, content links, even just how you doing? notes to build a relationship.
Pretty sweet – AND effective.
https://resultleadgeneration.com to discover what Web Visitors Into Leads can do for your business.
You could be converting up to 100X more leads today!
Eric
PS: Web Visitors Into Leads offers a complimentary 14-day trial – and it even includes International Long Distance Calling.
You have customers waiting to talk with you right now… don’t keep them waiting.
https://resultleadgeneration.com to try Web Visitors Into Leads now.
If you’d like to Want to receive fewer emails, or none whatsoever? Update your email preferences by visiting https://resultleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hello Lucofi Owner!
My name’s Eric and I just came across your website at Lucofi…
I found it after a quick search, so your SEO’s working out…
Content looks pretty good…
One thing’s missing though…
A QUICK, EASY way to connect with you NOW.
Because studies show that a web lead like me will only hang out a few seconds – 7 out of 10 disappear almost instantly, Surf Surf Surf… then gone forever.
I have the solution:
Web Visitors Into Leads is a software widget that works on your site, ready to capture any visitor’s Name, Email address, and Phone Number. You’ll know immediately they’re interested and you can call them directly to TALK with them – literally while they’re still on the web looking at your site.
https://resultleadgeneration.com to try out a Live Demo with Web Visitors Into Leads now to see exactly how it works and even give it a try… it could be huge for your business.
When targeting leads, you HAVE to act fast – the difference between contacting someone within 5 minutes versus 30 minutes later can be huge – like 100 times better!
That’s why you should check out our new SMS Text With Lead feature as well… once you’ve captured the phone number of the website visitor, you can automatically kick off a text (SMS) conversation with them.
Imagine how powerful this could be – even if they don’t take you up on your offer immediately, you can stay in touch with them using text messages to make new offers, provide links to great content, and build your credibility.
Just this alone could be a game changer to make your website even more effective.
Strike when the iron’s hot!
https://resultleadgeneration.com to discover what Web Visitors Into Leads can do for your business, potentially converting up to 100X more eyeballs into leads today!
Eric
PS: Web Visitors Into Leads offers a complimentary 14-day trial – you could be converting up to 100x more leads immediately!
It even includes International Long Distance Calling.
You have customers waiting to talk with you right now… don’t keep them waiting.
https://resultleadgeneration.com to try Web Visitors Into Leads now.
If you’d like to Want to receive fewer emails, or none whatsoever? Update your email preferences by visiting https://resultleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hello to the Lucofi Owner,
This is Eric here, with a brief note about your website Lucofi.
I am on the internet often, and I see many business websites.
Similar to yours, several of them have excellent content.
However, too often, they fall short when it comes to interacting and connecting with a visitor.
I understand – it is challenging. Research shows that many individuals who arrive at a site leave within moments without sharing any details. You gained their initial attention, but nothing else.
Here is a possible solution:
Web Visitors Into Leads is a tool that operates on your site, prepared to collect each visitor’s name, email address, and phone number. You will know right away that they are interested, and you could speak with them while they are still online viewing your site.
Please visit https://actionleadgeneration.com to view a Live Demo with Web Visitors Into Leads today to see exactly how it operates.
It can be significant for your business – and because you have their phone number, with our SMS Text With Lead capability, you can instantly begin a text conversation. Connecting with someone in those first minutes is far more effective than waiting longer.
Additionally, with text messaging, you may follow up later with new updates, content links, or simple notes to keep the conversation moving.
Everything described is straightforward to implement, cost-effective, and genuinely helpful.
Please visit https://actionleadgeneration.com to learn what Web Visitors Into Leads can provide for your business.
You can be engaging with more visitors as we speak!
Eric
P.S. Web Visitors Into Leads provides a 14-day evaluation – and it includes international calling. You have individuals who may be ready to speak with you now, so please do not delay.
Please visit https://actionleadgeneration.com to view Web Visitors Into Leads today.
If you’d like to Want to receive fewer emails, or none whatsoever? Update your email preferences by visiting https://actionleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hi Lucofi Owner!
My name’s Eric and I’m betting you’d like your website Lucofi to generate more leads.
Here’s how:
LeadConnect is a software widget that works on your site, ready to capture any visitor’s Name, Email address, and Phone Number. It signals you as soon as they say they’re interested – so that you can talk to that lead while they’re still there at Lucofi.
https://blastleadgeneration.com for a live demo now.
Plus, now that you’ve got their phone number, our new SMS Text With Lead feature enables you to start a text (SMS) conversation – answer questions, provide more info, and close a deal that way.
If they don’t take you up on your offer then, just follow up with text messages for new offers, content links, even just “how you doing?” notes to build a relationship.
https://blastleadgeneration.com to discover what LeadConnect can do for your business.
The difference between contacting someone within 5 minutes versus a half-hour means you could be converting up to 100X more leads today!
Try LeadConnect and get more leads now.
Eric
PS: The studies show 7 out of 10 visitors don’t hang around – you can’t afford to lose them!
LeadConnect offers a complimentary 14-day trial – and it even includes International Long Distance Calling.
You have customers waiting to talk with you right now… don’t keep them waiting.
https://blastleadgeneration.com to try LeadConnect now.
If you’d like to Want to receive fewer emails, or none whatsoever? Update your email preferences by visiting https://blastleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hello Lucofi
I just found your site, quick question…
My name’s Eric, and I just found your site – Lucofi – while surfing the net. You showed up at the top of the search results, so I checked you out. Looks like what you’re doing is pretty cool.
But if you don’t mind me asking – after someone like me stumbles across Lucofi, what usually happens?
Is your site generating leads for your business?
I’m guessing some, but I also bet you’d like more… research indicates that 7 out of 10 who land on a site wind up leaving without a trace.
Not good.
Here’s an idea…
How about making it really EASY for every visitor who shows up to get a personal phone call from you as soon as they hit your site…
You can –
LeadConnect is a software widget that works on your site, ready to capture any visitor’s Name, Email address and Phone Number. It signals you the moment they let you know they’re interested – so that you can talk to that lead while they’re literally looking over your site.
https://blastleadgeneration.com to try out a Live Demo with LeadConnect now to see exactly how it works.
You’ll be amazed – the difference between contacting someone within 5 minutes versus 30 minutes later could boost your results 100-fold.
It gets even better… once you’ve captured their phone number, with our new SMS Text With Lead feature, you can instantly start a text (SMS) conversation.
That way, even if you don’t close a deal right away, you can follow up with text messages for new offers, content links, even just, how you doing? notes to build a relationship.
Pretty sweet – AND effective.
https://blastleadgeneration.com to discover what LeadConnect can do for your business.
You could be converting up to 100X more leads today!
Eric
PS: LeadConnect offers a FREE 14 days trial – you could be converting up to 100x more leads immediately!
It even includes International Long Distance Calling.
You have customers waiting to talk with you right now… don’t keep them waiting.
https://blastleadgeneration.com to try LeadConnect now.
If you’d like to Want to receive less emails, or none whatsoever? Update your email preferences by visiting https://blastleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hello to the Lucofi Owner,
I am Eric, and I recently discovered your site, Lucofi.
Your site has a great deal going for it, but I have an idea to help make it even more effective.
Please take a look at Web Visitors Into Leads – you can visit https://actionleadgeneration.com for a demonstration today.
Web Visitors Into Leads is a tool that operates on your site, prepared to gather each visitor’s name, email address, and phone number. It notifies you the instant they show interest, allowing you to speak with them while they are still viewing your site.
Once you have their phone number, you can begin an SMS conversation immediately. If they are not ready at that moment, you can follow up later with messages to share updates, information, or simply check in and maintain a connection.
Please visit https://actionleadgeneration.com to learn more about what Web Visitors Into Leads can do for your business.
Connecting with someone promptly rather than waiting a long time can make a substantial difference in your results.
Eric
P.S. Research indicates that many site visitors leave quickly without returning. Web Visitors Into Leads includes a 14-day no-cost trial and supports international communication. There may be individuals ready to engage with you right now, so please do not delay.
Please visit https://actionleadgeneration.com to learn more about Web Visitors Into Leads.
If you’d like to Want to receive fewer emails, or none whatsoever? Update your email preferences by visiting https://actionleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hello Lucofi Owner,
My name’s Eric and for just a second, imagine this:
– Someone does a search and ends up at Lucofi.
– They hang out for a moment to check it out. I’m interested… but… maybe…
– And then they click the back button and check out the other search results instead.
– Bottom line – you got an eyeball, but nothing else.
– They’re gone.
This isn’t really your fault – it happens a lot – studies show 7 out of 10 visitors to any site leave without leaving a trace.
But you can fix that.
Web Visitors Into Leads is a software widget that works on your site, ready to gather any visitor’s Name, Email address, and Phone Number. It lets you know immediately – allowing you to call that lead while they’re literally looking over your site.
Please see this URL to test a Live Demo with Web Visitors Into Leads now to see exactly how it works:
https://trustedleadgeneration.com
Time is of the essence when it comes to connecting with leads – the difference between contacting someone within 5 minutes compared to 30 minutes later can be significant.
Additionally, now that you have their phone number, with our new SMS Text With Lead feature you can immediately start a text conversation… so even if you don’t close a deal then, you can follow up with text messages for new offers, useful links, even just a quick note to build a relationship.
Effective stuff.
Please see this URL to learn what Web Visitors Into Leads can do for your business:
https://trustedleadgeneration.com
You could be converting significantly more leads today!
Eric
PS: Web Visitors Into Leads offers a 14-day evaluation – it even includes international calling.
You have customers ready to talk with you right now… don’t keep them waiting.
Please see this URL to try Web Visitors Into Leads now:
https://trustedleadgeneration.com
If you’d like to Want to receive less emails, or none whatsoever? Update your email preferences by clicking here. click here https://trustedleadgeneration.com/unsubscribe.aspx?d=lucofi.com
Hello to the Lucofi Manager,
I am Eric and I’m thinking you would like your website Lucofi to produce additional inquiries.
Here is one approach:
Web Visitors Into Leads is a tool that operates on your site, prepared to capture each visitor’s name, email, and phone number. It informs you as soon as someone expresses interest, allowing you to speak with them while they are still viewing Lucofi.
View a Demonstration https://actionleadgeneration.com to see how Web Visitors Into Leads works.
Once you have their phone number, our new SMS Text With Lead feature allows you to start a text conversation. You can answer questions, provide more details, and even finalize a connection that way.
If they are not ready at that moment, you can follow up later with text messages that share updated information, helpful links, or simply keep the dialogue open to establish a positive rapport.
Learn More https://actionleadgeneration.com to understand how Web Visitors Into Leads can support your business, potentially engaging many more interested individuals.
Eric
P.S. Studies suggest that most visitors leave quickly. This resource offers a 14-day evaluation period and also includes international communication support. There may be individuals ready to speak with you now, so please don’t let the opportunity pass.
If you’d like to Want to receive fewer emails, or none whatsoever? Update your email preferences by visiting https://actionleadgeneration.com/unsubscribe.aspx?d=lucofi.com